10.7 转录组单组学快速分析示例
本章节将利用示例数据,完成一个转录组单组学的从导入的完成基本常见的分析流程。
示例数据下载:1. Github下载地址 2. 百度网盘下载地址
10.7.1 转录组数据导入
注意:
由于单一组学项目不涉及创建关系表,因此表达矩阵的样本名称必须与表型数据中样本名称完全一致。
由于单一组学项目不涉及创建关系表,因此表达矩阵的样本名称必须与表型数据中样本名称完全一致。
library(EasyMultiProfiler)
meta_data <- read.table('col.txt',header = T,row.names = 1)
data <- read.table('rna.txt',header = T,sep = '\t')
data <- data[!duplicated(data[,1]),] # 去除因为excel导致的部分基因名称重复
MAE <- EMP_easy_import(data = data,coldata = meta_data,type = 'normal')
10.7.2 转录组数据查看
查看当前转录组组学
MAE |>
EMP_assay_extract() # 查看表达矩阵
MAE |>
EMP_coldata_extract() # 查看表型数据
MAE |>
EMP_rowdata_extract() # 查看基因注释
10.7.3 基因丰度批次矫正 (非必须)
按照表型数据中采集地的因素进行矫正
MAE |>
EMP_assay_extract() |>
EMP_adjust_abundance(.factor_unwanted = 'Region',
.factor_of_interest = 'Group',
method = 'combat_seq')
10.7.4 基因ID的自由转换
EMP包内置了Human, Mouse,Pig和Zebrafish的基因注释集。 将SYMBOL转换成ENTREZID
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',species = 'Human')
将SYMBOL转换成ENSEMBL
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENSEMBL',species = 'Human')
如果内置基因集没有所需物种,也可以使用orgdb系列包进行转换
library(org.Hs.eg.db)
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENSEMBL',OrgDb = org.Hs.eg.db)
10.7.5 增加基因-疾病的相关注释
增加基因相关疾病注释,目前仅支持Human_disease和Mouse_disease
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',add ='Human_disease') |>
EMP_assay_extract(pattern = 'cancer',pattern_ref = 'Human_disease')
10.7.6 基因丰度转换
MAE |>
EMP_assay_extract() |>
EMP_decostand(method = 'log2+1')
10.7.7 核心基因筛选 (非必须)
利用edgeR内置算法进行核心基因集的筛选
MAE |>
EMP_assay_extract() |>
EMP_identify_assay(method = 'edgeR',
min = 10,min_ratio = 0.7)
10.7.8 差异基因筛选
利用DESeq算法进行差异分析,并绘制火山图
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group) |>
EMP_volcanol_plot(show='pic',key_feature = c('CCL20','ZBTB7C'),
palette = c('#FA7F6F','#96C47D','#BEB8DC'),
dot_size = 2.5,threshold_x = 1,mytheme = "theme_light()",
min.segment.length = 0, seed = 42, box.padding = 0.5)
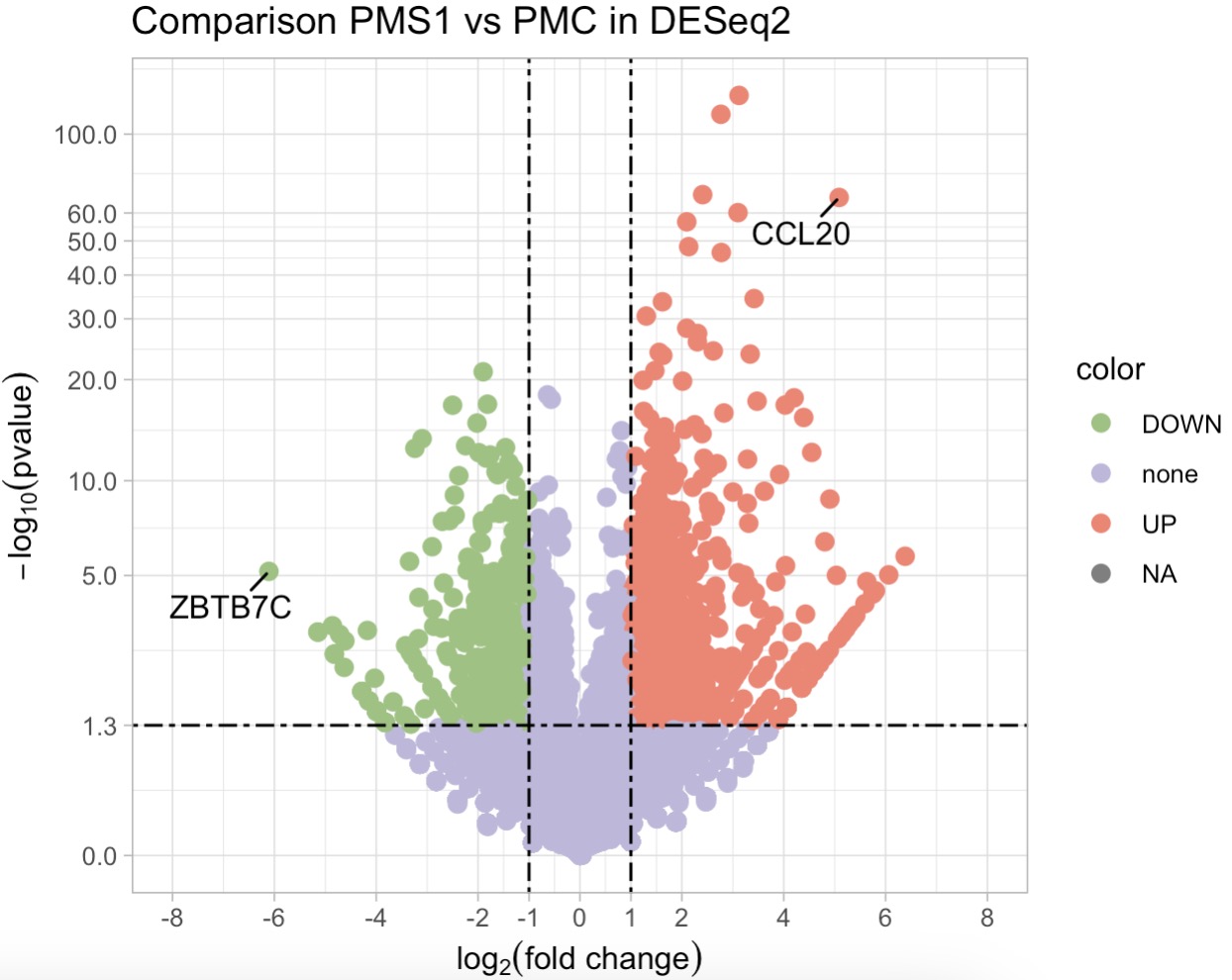
更多差异分析算法,并筛选出差异显著基因
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'edgeR_quasi_likelihood',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = TRUE)
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'edgeR_likelihood_ratio',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = TRUE)
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'edger_robust_likelihood_ratio',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = TRUE)
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'limma_voom',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = TRUE)
MAE |>
EMP_assay_extract() |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'limma_voom_sample_weights',.formula = ~Group) |>
EMP_filter(feature_condition = pvalue < 0.05,keep_result = TRUE)
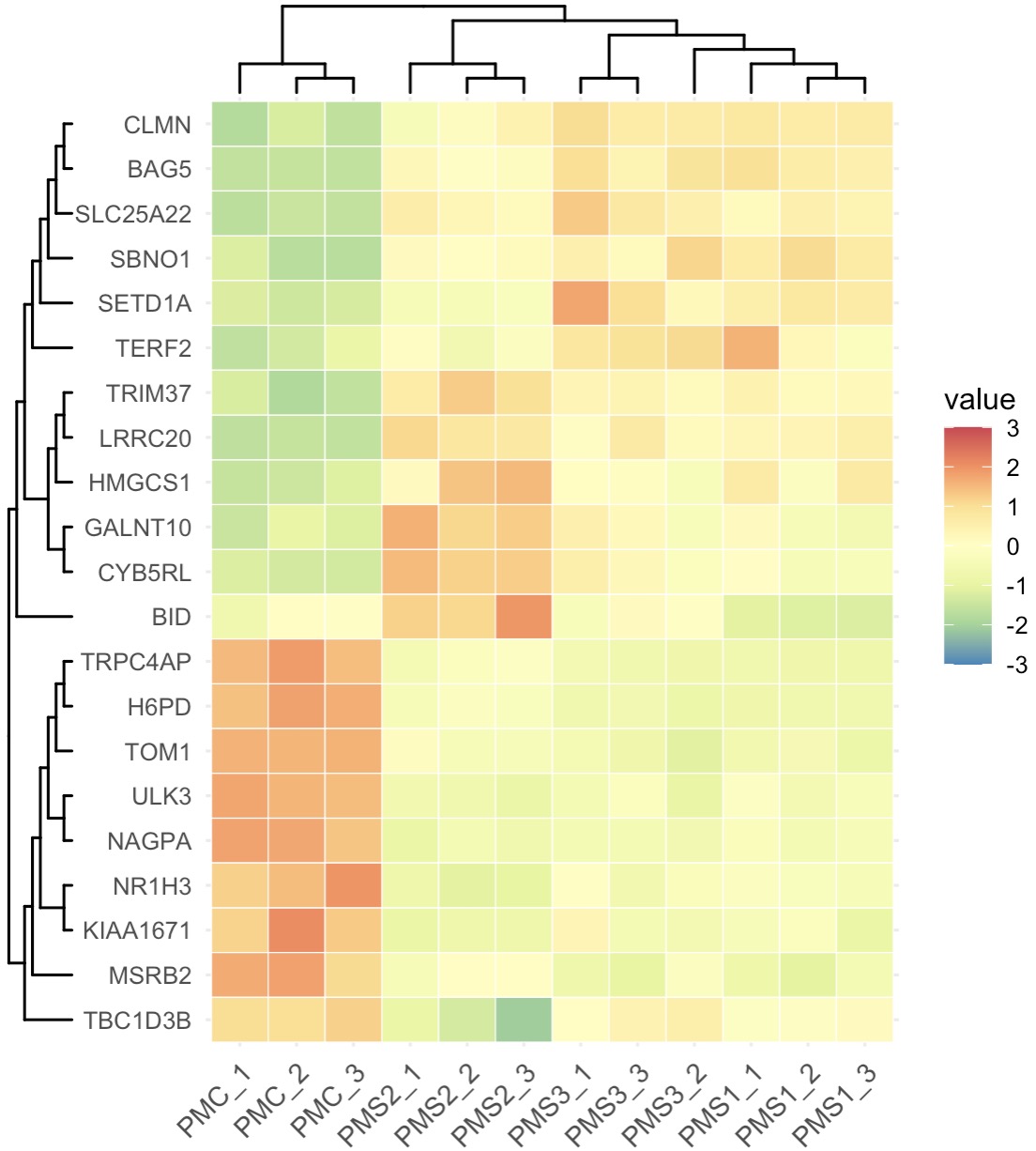
10.7.9 机器学习基因筛选
EMP包内置了Boruta算法、随机森林算法、xgboost算法和Lasso算法进行特征筛选。 详细用法可以使用help(EMP_marker_analysis)查看更多示例。
利用Boruta算法快速进行筛选
MAE |>
EMP_assay_extract() |>
EMP_marker_analysis(method = 'boruta',estimate_group = 'Group') |>
EMP_filter(feature_condition = Boruta_decision!= 'Rejected') |>
EMP_heatmap_plot(palette='Spectral',legend_bar='auto',
scale='standardize',
clust_row=TRUE,clust_col=TRUE)
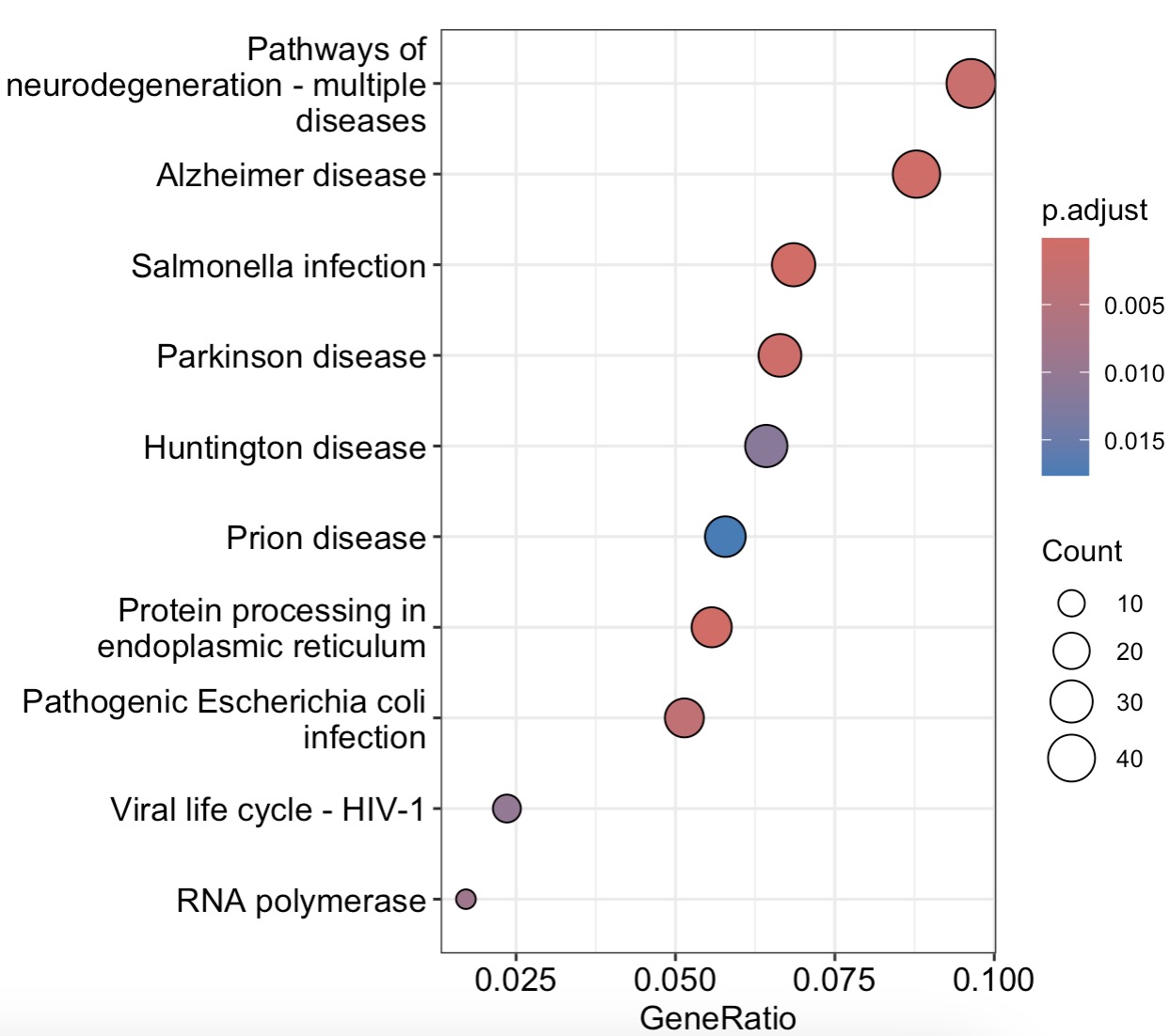
10.7.10 ORA富集
options(timeout = 180) # 由于连接数据库需要时间,建议增加这行避免数据拉取失
基于KEGG的富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',
species = 'Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method='DESeq2',.formula = ~Group) |>
EMP_enrich_analysis( pvalue<0.05,keyType ='entrezid',
KEGG_Type = 'KEGG',species='hsa',
pvalueCutoff=0.05) |>
EMP_enrich_dotplot()
基于GO的富集
library(org.Hs.eg.db)
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'symbol',to='entrezid',species='Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group,
p.adjust = 'fdr') |>
EMP_enrich_analysis(pvalue<0.05,method = 'go',OrgDb=org.Hs.eg.db,
ont='MF',readable=TRUE,pvalueCutoff=0.05) |>
EMP_enrich_dotplot(show=6)
基于DOSE的富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'symbol',to='entrezid',
species='Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'DESeq2',
.formula = ~Group,p.adjust = 'fdr') |>
EMP_enrich_analysis(pvalue<0.05,method = 'do',ont="HDO",
organism= 'hsa',readable=TRUE,
pvalueCutoff=0.05) |>
EMP_enrich_dotplot(show=5)
基于Reactome的富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'symbol',to='entrezid',
species='Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method = 'DESeq2',.formula = ~Group,
p.adjust = 'fdr') |>
EMP_enrich_analysis(pvalue<0.05,method = 'Reactome',organism= 'human',
readable=TRUE,pvalueCutoff=0.05) |>
EMP_enrich_dotplot()
10.7.11 GSEA富集
options(timeout = 180) # 由于连接数据库需要时间,建议增加这行避免数据拉取失
GSEA有三种排序方法,下方示例仅在KEGG数据库进行示范。
用户可根据ORA富集的参数设置,在Go、DOSE和Reactome进行GSEA富集。
基于信噪比排序方法的KEGG富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',species = 'Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_GSEA_analysis(pvalue<0.05,method='signal2Noise',
estimate_group = 'Group',species = 'hsa',
pvalueCutoff = 1,keyType = 'entrezid') |>
EMP_GSEA_plot(geneSetID='hsa04930')
基于差异分析排序方法的KEGG富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',
species = 'Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_diff_analysis(method='DESeq2',.formula = ~0+Group,
group_level=c('PMC','PMS1')) |>
EMP_GSEA_analysis(method='log2FC',enrich_method = 'kegg',
species = 'hsa',keyType = 'entrezid',
pvalueCutoff = 0.05) |>
EMP_GSEA_plot(geneSetID='hsa04950')
基于相关分析排序方法的KEGG富集
MAE |>
EMP_assay_extract() |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',
species = 'Human') |>
EMP_filter(Group %in% c('PMC','PMS1')) |>
EMP_GSEA_analysis(method='cor',enrich_method = 'kegg',
keyType='entrezid',estimate_group = 'NR5A2',
cor_method = 'spearman',
pvalueCutoff = 0.05,species='hsa') |>
EMP_GSEA_plot(geneSetID='hsa05415')
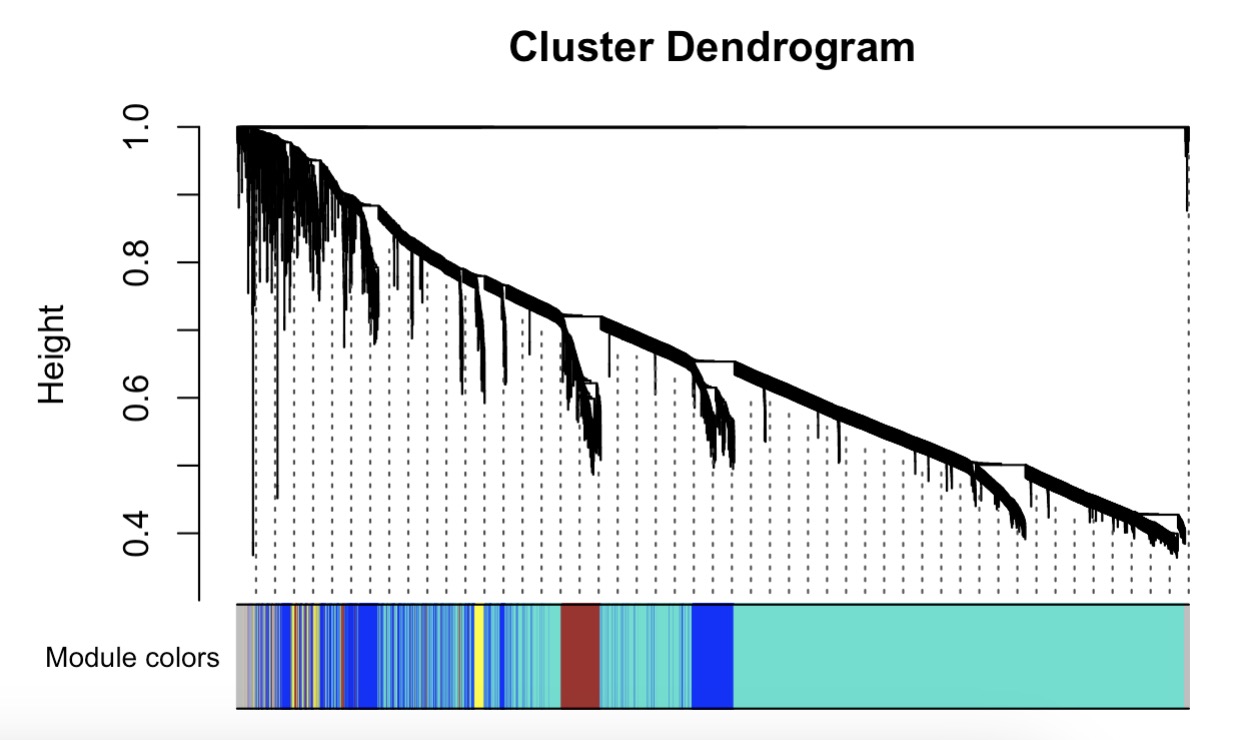
10.7.12 WGCNA分析
第一步:根据表型数据获取聚类情况
MAE |>
EMP_assay_extract() |>
EMP_identify_assay(method = 'edgeR',estimate_group = 'Group') |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',species = 'Human') |>
EMP_WGCNA_cluster_analysis()
第二步:根据表型数据获取相关基因簇热图
MAE |>
EMP_assay_extract() |>
EMP_identify_assay(method = 'edgeR',estimate_group = 'Group') |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',species = 'Human') |>
EMP_WGCNA_cluster_analysis() |>
EMP_WGCNA_cor_analysis(coldata_to_assay = c('NR5A2','HNF4G','HNF1B','PAX4','RFX6','NEUROG3'),
method='spearman') |>
EMP_heatmap_plot(palette = 'Spectral')
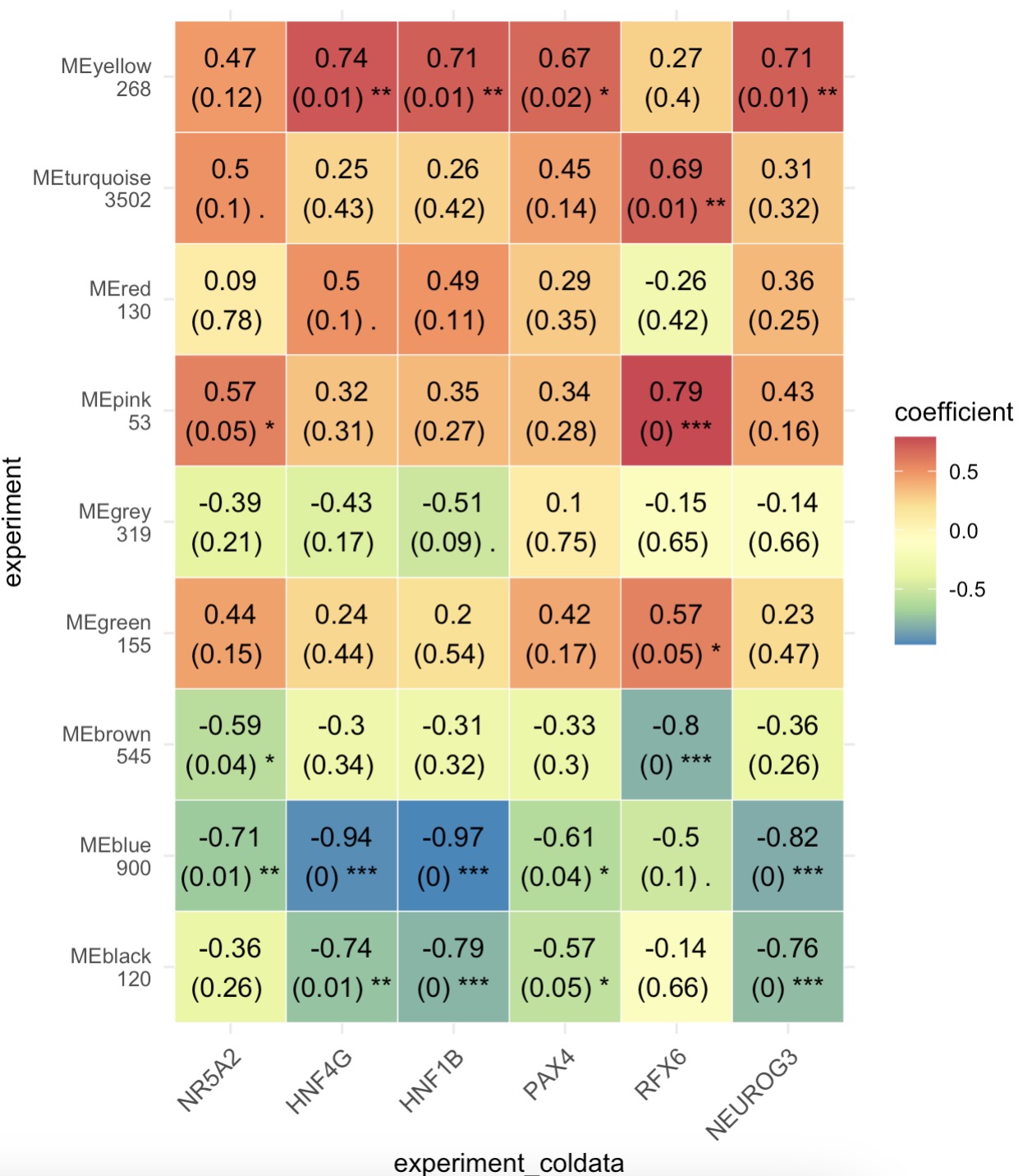
第三步:筛选相关基因簇进行富集分析
MAE |>
EMP_assay_extract() |>
EMP_identify_assay(method = 'edgeR',estimate_group = 'Group') |>
EMP_feature_convert(from = 'SYMBOL',to = 'ENTREZID',species = 'Human') |>
EMP_WGCNA_cluster_analysis() |>
EMP_WGCNA_cor_analysis(coldata_to_assay = c('NR5A2','HNF4G','HNF1B','PAX4','RFX6','NEUROG3'),
method='spearman') |>
EMP_heatmap_plot(palette = 'Spectral') |>
EMP_filter(feature_condition = WGCNA_color == 'blue' ) |>
EMP_enrich_analysis(keyType = 'entrezid',species = 'hsa') |>
EMP_enrich_dotplot()